Research Prof. Dr. Dina Grohmann
Our Vision
Heterogeneity is a fundamental property in biology. We strive to identify and describe biological heterogeneity and diversity on the organismic and molecular level and aim to understand how biological heterogeneity drives the function of molecular machineries.
On the organismic level, we are committed to expand our understanding of microbial diversity on our planet. Having isolated more than 1.000 archaeal strains from all over the world over the last three decades, we are especially intrigued by organisms from the third domain of life, Archaea, and are home of the Archaea Centre Regensburg. In addition, we also look on the evolutionary journey of fundamental biological machineries and molecular processes that are present in all living cells. Here, we focus on processes along the gene expression pathway including the regulation of gene expression at the transcriptional (transcription factors and transcription machinery) and posttranscriptional level (RNA processing, RNA-guided interference and archaeal Sm like proteins) as well as prokaryotic defence system (CRISPR-Cas and prokaroytic Argonaute).
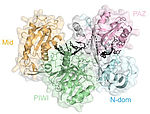
Molecular mechanisms of Argonaute proteins
[D. Grohmann / S. Willkomm]
Molecular mechanisms of Argonaute proteins
Despite the mechanistic and structural similarities between archaeal, bacterial and eukaryotic Argonaute proteins, the biological function of prokaryotic Argonaute proteins is only poorly understood. We aim to unravel the biological function of prokaryotic Argonautes. Furthermore, we are especially interested in the structural features and dynamic behaviour of Argonaute that promote substrate recognition and cleavage, thereby revealing differences and similarities in eukaryotic and prokaryotic Argonaute biology.
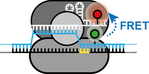
Single-molecule analysis of CRISPR-Cas enzymes
Single-molecule analysis of CRISPR-Cas enzymes
CRISPR-Cas systems constitute the adaptive immune systems of bacteria and archaea. As we scientist only discoverd CRISPR-Cas a few years ago, there is much to be learned about the diversity and function of these complex defence systems. Employing biophysical technqiues like single-molecule FRET, we seek to understand the molecular mechanisms and conformational dynamics of a subset of Cas proteins.
Isolation and characterization of new archaeal species

[R. Reichelt / D. Grohmann / R. Rachel]
Isolation and characterization of new archaeal species
Since several decades, the investigation of microbial populations in extreme biotopes is one of our major research fields. Besides sampling of continental and marine high temperature biotopes (e.g. black smokers) we also hunt in habitats with moderate temperatures for new archaeal strains. After their enrichment and isolation, we concentrate on the physiological and molecular characterization but also on an approach for biotechnological application of the novel microorganisms. Among others, we currently focus on novel consortia from low temperature biotopes in Costa Rica, which consist of different archaeal and bacterial representatives.
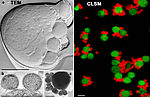
Characterisation of the Ignicoccus-Nanoarchaeum interaction/symbiosis
[D. Grohmann]
Characterisation of the Ignicoccus-Nanoarchaeum interaction/symbiosis
The intimate association of Ignicoccus hospitalis and Nanoarchaeum equitans is a microbial community solely formed by archaeal cells. This fascinating symbiosis was first isolated and cultivated under laboratory conditions in the Institute of Microbiology at the University of Regensburg. While I. hospitalis can be grown also in pure culture, proliferation of N. equitans cells is strictly dependent on an assocation with I. hospitalis cells. N. equitans is one of the smallest living cells known. We are studying this intriguing archaeal community to answer question that concern the intermembrane system and energy production of how N. equitans can thrive as a minimalistic cell.
Regulatory networks in Archaea
[W. Hausner]
Regulatory networks in Archaea
The genome of Pyrococcus furiosus encodes 85 putative transcription factors but the function of most of these proteins and the corresponding gene regulatory networks are unknown. To unravel the function of some of these proteins we combine traditional biochemical in vitro techniques to identify binding sequences and transcription profiles of individual genes with global approaches like chromatin immunoprecipitation and RNA sequencing to reconstruct gene regulatory networks. Our latest project deals with CopR, a transcription regulator which is necessary to survive at elevated copper concentrations.
Studying the archaeal model organism Pyrococcus furiosus
[W. Hausner / D. Grohmann]
Studying the archaeal model organism Pyrococcus furiosus
Pyrococcus furiosus was isolated from Vulcano island in Italy and described more than 30 years ago at our Institute of Microbiology in Regensburg. With an optimal growth temperature near the boiling point of water and a doubling time of ~37 min it serves as a model organism to explore the biology of hyperthermophiles. To enable a more detailed functional analysis we have established a genetic system allowing plasmid-based expression of proteins as well as markerless deletion of individual genes. In our recent work, we provide detailed sequence information on the DNA and the RNA level by using state-of-the-art sequencing techniques (Illumina, PacBio and Nanopore sequencing). An update on the protein level using MALDI-TOF mass spectrometry analysis is in progress.
Exploiting methanogenic archaea for Power-to-Gas applications
[A. Bellack]
Exploiting methanogenic archaea for Power-to-Gas applications
Power-to-Gas is an innovative concept to convert surplus electricity from renewable energy sources to hydrogen either for direct usage or for reduction of carbon dioxide to methane. The biologically or chemically produced methane is of special interest since it can be fed into the existing gas network for storage and distribution. In the joint project ‘ORBIT’, we aim to optimize a trickle-bed reactor for biological methanation in collaboration with process technicians, power engineers, and field-leading industrial partners. Central part of the project is the analysis of different combinations of methanogenic archaea and reactor packings to increase the methane production rate and methane yield of the process.