


EpiGenetics
Chromatin Dynamics and Nuclear Architecture
Prof. Gernot Längst - Biochemistry III
Lab activities
Collaborative Research Center and Transregio projects
Startup Company
Bioinformatics
Graduate School
Recent News
Corona: Mit High-Tech gegen das Virus. - Link - zur Webseite des Bayerischen Rundfunks.
- Link - zum Bericht in der Abenschau vom 23.2.2021
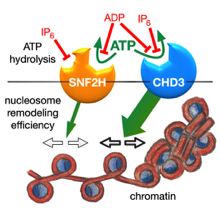
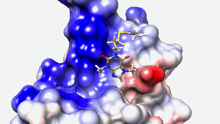
Sequence and functional differences in the ATPase domains of CHD3 and SNF2H promise potential for selective regulability and drugability. Helen Hoffmeister et al. FEBS J. 2021, doi: 10.1111/febs.15699
Chromatin remodelers use the energy of ATP hydrolysis to regulate chromatin dynamics. Their impact for development and disease requires strict enzymatic control. Here, we address the differential regulability of the ATPase domain of hSNF2H and hCHD3, exhibiting similar substrate affinities and enzymatic activities. Both enzymes are comparably strongly inhibited in their ATP hydrolysis activity by the competitive...

Targeting Chromatin Remodeling to inhibit Tumor Metastasis
Chromatin remodeling complexes are ATP-dependent enzymes that regulate all DNA dependent processes by moving, exchanging orevicting nucleosomes from the underlying DNA. These epigenetic modifiers play important roles in cellular homeostasis and are preferentially mutated in many diseases and cancer. Baz2a is aremodeling protein that is targeted to the genome by non-coding RNAsand highly upregulated in breast cancer. We aim to screen for small molecule inhibitors of Baz2a RNA interaction to block its tumorigenic potential and to evaluate these in a...

Die Verpackung des SARS-CoV-2 Genoms in das Viruspartikel ist ein zentraler Schritt derVirusvermehrung, der durch das virale N-Protein reguliert wird. Im Verbund von Biochemikern,Zellbiologen, Drug-Discovery-Experten und Virologen werden wir Inhibitoren der N-Protein-Funktionidentifizieren. Die Unterbindung der Genomverpackung hat das Ziel, die Bildung infektiöser Viren zuunterbinden und den viralen Lebenszyklus zu unterbrechen.
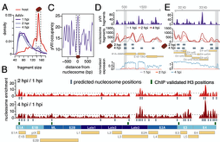
A pVII positioning code determines the dynamics of Adenoviral nucleoprotein structure and primes it for early gene activation Uwe Schwartz et al., (2020) BioRxiv; ID: https://doi.org/10.1101/2020.07.07.190926
Inside the capsid adenovirus DNA is associated with the structural protein pVII. However, the viral DNA organisation, pVII positioning and the dynamic changes of viral packaging upon infection, to form a transcriptional active genome, are not known. We combined MNase-Seq and single genome imaging during early infection to provide the structure and time resolved dynamics of viral chromatin changes, correlated...
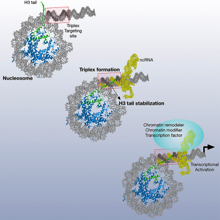
Nucleosomes Stabilize ssRNA-dsDNA Triple Helices in Human Cells Rodrigo Maldonado et al., (2019) Molecular Cell; PMID: 30770238
Chromatin-associated non-coding RNAs modulate the epigenetic landscape and its associated gene expression program. The formation of triple helices is one mechanism of sequence-specific targeting of RNA to chromatin. With this study, we show an important role of the nucleosome and its relative positioning to the triplex targeting site (TTS) in stabilizing RNA-DNA triplexes in vitro and in vivo. Triplex stabilization depends on the histone H3 tail and the location of the TTS close...
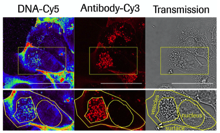
Antibody-targeted chromatin enables effective intracellular delivery and functionality of CRISPR/Cas9 expression plasmids Killian et al., (2019) Nucleic Acids Research; PMID: 30809660
We report a novel system for efficient and specific targeted delivery of large nucleic acids to and into cells. Plasmid DNA and core histones were assembled to chromatin by salt gradient dialysis and subsequently connected to bispecific antibody derivatives (bsAbs) via a nucleic acid binding peptide bridge. The resulting reconstituted vehicles termed ‘plasmid-chromatin’ deliver packaged nucleic acids to and into cells expressing...
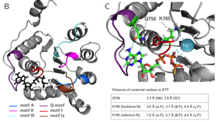
Elucidation of the functional role of the Q- and I-motif in the human chromatin remodeling enzyme BRG1
Helen Hoffmeister et al., (2019) JBC; PMID: 30647132
The Snf2 proteins, comprising 53 different enzymesin humans, belong to the SF2 family. Many Snf2enzymes possess chromatin remodeling activity,requiring a functional ATPase domain consisting ofconserved motifs named Q and I-VII. These motifsform two recA-like domains, creating an ATP-binding pocket. Little is known about the functionof the conserved motifs in chromatin remodelingenzymes. Here, we characterized the function of theQ- and I (Walker I)-motif...

Characterizing the nuclease accessibility of DNA in human cells to map higher order structures of chromatin.
Uwe Schwartz et al., (2018) Nucleic Acids Research; PMID: 30496478
Packaging of DNA into chromatin regulates DNA ac-cessibility and consequently all DNA-dependent pro-cesses. The nucleosome is the basic packaging unitof DNA forming arrays that are suggested, by bio-chemical studies, to fold hierarchically into orderedhigher-order structures of chromatin. This organiza-tion has been recently questioned using microscopytechniques, proposing an irregular structure. To ad-dress the principles of chromatin...
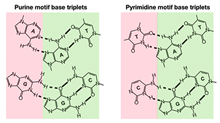
Rodrigo Maldonado et al., (2017) RNA; PMID: 29222118
Triplexes are noncanonical DNA structures, which are functionally associated with regulation of gene expression through ncRNAtargeting to chromatin. Based on the rules of Hoogsteen base-pairing, polypurine sequences of a duplex can potentially formtriplex structures with single-stranded oligonucleotides. Prediction of triplex-forming sequences by bioinformatics analyseshave revealed enrichment of potential triplex targeting sites (TTS) at regulatory elements, mainly in promoters and enhancers,suggesting a potential function of RNA–DNA triplexes in transcriptional...
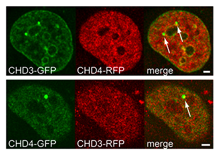
Helen Hoffmeister et al. (2017) Nucleic Acids Research; PMID: 28977666
CHD3 and CHD4 (Chromodomain Helicase DNA bind-ing protein), two highly similar representatives of theMi-2 subfamily of SF2 helicases, are coexpressed inmany cell lines and tissues and have been reportedto act as the motor subunit of the NuRD complex(nucleosome remodeling and deacetylase activities).Besides CHD proteins, NuRD contains several re-pressors like HDAC1/2, MTA2/3 and MBD2/3, argu-ing for a role as a transcriptional repressor. However,the subunit composition varies among cell- and tis-sue types and physiological conditions. In...
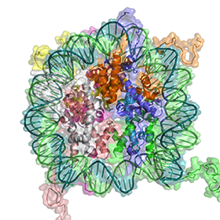
Elisabeth Silberhorn et al., (2016) Plos Pathogen; PMID: 28033404
Nucleosomes are not positioned randomly on DNA but on preferential sites with respect to the underlying DNA sequence. Histones belong to the most conserved eukaryotic proteins, as sequence dependent nucleosome positioning is an essential regulatory feature of nucleosomes, determining the accessibility of regulatory factors to DNA. We determined the biochemical properties of plasmodium histones and show that they are distinct from human forms, explaining the accessible chromatin structure of falciparum. Amino acid exchanges in the histones do...
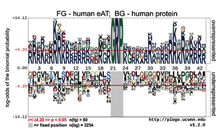
Michael Filarsky et al., RNA Biol. 2015;12(8):864-76
The AT-hook has been defined as a DNA binding peptide motif that contains a glycine-arginine-proline (G-R-P) tripeptide core flanked by basic amino acids. Recent reports documented variations in the sequence of AT-hooks and revealed RNA binding activity of some canonical AT-hooks, suggesting a higher structural and functional variability of this protein domain than previously anticipated. Here we describe the discovery and characterization of the extended AT-hook peptide motif (eAT-hook), in which basic amino acids appear symmetrical mainly at a distance...
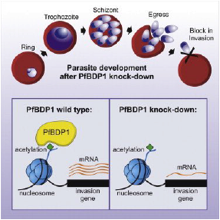
Josling et al., Cell Host Microbe. 2015 Jun 10;17(6):741-51
During red-blood-cell-stage infection of Plasmodium falciparum, the parasite undergoes repeated rounds of replication, egress, and invasion. Erythrocyte invasion involves specific interactions between host cell receptors and parasite ligands and coordinated expression of genes specific to this step of the life cycle. We show that a parasite-specific bromodomain protein, PfBDP1, binds to chromatin at transcriptional start sites of invasion-related genes and directly controls their expression. Conditional PfBDP1 knockdown causes a dramatic defect in...
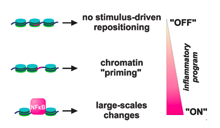
Sarah Diermeier et al., Genome Biology 2014
Background: The rearrangement of nucleosomes along the DNA fiber profoundly affects gene expression, but little is known about how signalling reshapes the chromatin landscape, in three-dimensional space and over time, to allow establishment of new transcriptional programs.
Results: Using micrococcal nuclease treatment and high-throughput sequencing, we map genome-wide changes in nucleosome positioning in primary human endothelial cells stimulated with tumour necrosis factor alpha (TNFα) - a proinflammatory cytokine that signals through nuclear factor kappa-B...



EpiGenetics
Chromatin Dynamics and Nuclear Architecture
Prof. Gernot Längst - Biochemistry III
Lab activities
Collaborative Research Center and Transregio projects
Startup Company
Bioinformatics
Graduate School
Recent News